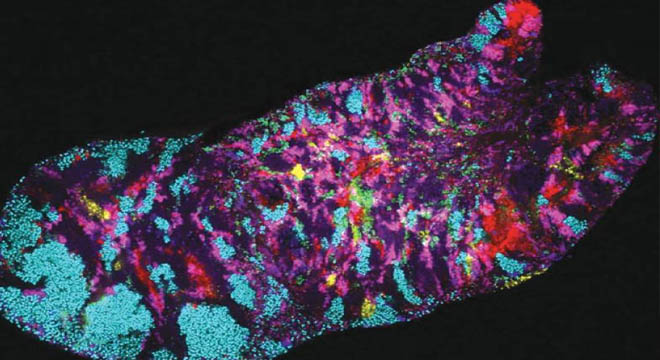
ISLAMABAD-Researchers take a closer look at the genomes of microbial communities in the human mouth.Bacteria often show very strong biogeography — some bacteria are abundant in specific locations while absent from others — leading to major questions when applying microbiology to therapeutics or probiotics: how did the bacteria get into the wrong place? How do we add the right bacteria into the right place when the biogeography has gotten ‘out of whack’?These questions, though, have one big obstacle, bacteria are so tiny and numerous with very diverse and complicated populations which creates major challenges to understanding which subgroups of bacteria live where and what genes or metabolic abilities allow them to thrive in these ‘wrong’ places.In a new study published in Genome Biology researchers led by Harvard University examined the human oral microbiome and discovered impressive variability in bacterial subpopulations living in certain areas of the mouth.Recent developments in sequencing and bioinformatic approaches have offered new ways to untangle the complexity of bacterial communities. Utter and Colleen Cavanaugh, Edward C. Jeffrey Professor of Biology in the Department of Organismic and Evolutionary Biology, Harvard University, teamed up with researchers at the Marine Biological Laboratory, Woods Hole, University of Chicago, and The Forsyth Institute to apply these state-of-the-art sequencing and analysis approaches to get a better picture of the oral microbiome.
Follow the PNI Facebook page for the latest news and updates.